So, last night’s analysis suggests that there’s not enough spread variability in the model. But it varied from run to run. So lets do a bunch of runs using replicate and see how far off we are.
n_init <- 50
Ler_params$gap_size <- 0
controls <- list(
n_reps = 10,
DS_seeds = TRUE,
ES_seeds = TRUE,
kernel_stoch = TRUE,
kernel_stoch_pots = TRUE,
seed_sampling = TRUE,
pot_width = 7
)The iteration and analysis, as a function to pass to replicate:
sim_mean_var <- function() {
Adults <- matrix(n_init, controls$n_reps, 1)
for (i in 1:6) {
Adults <- iterate_genotype(Adults, Ler_params, controls)
}
npot <- ncol(Adults)
rep_sum <- t(apply(Adults[, npot:1], 1, cummax))[, npot:1]
maxd <- apply(rep_sum, 1, function(x) max((1:length(x))[x > 0]))
maxd <- maxd[is.finite(maxd)]
result <- c(mean(maxd), var(maxd))
names(result) <- c("Mean", "Variance")
result
}Test the function:
sim_mean_var() Mean Variance
14.80000 13.73333 Do the replication
nruns <- 100
rep_spread_stats <- t(replicate(nruns, sim_mean_var(), simplify = TRUE))Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Infrep_spread_stats Mean Variance
[1,] 14.50000 6.7222222
[2,] 13.10000 5.4333333
[3,] 15.20000 7.2888889
[4,] 18.00000 74.6666667
[5,] 14.88889 3.3611111
[6,] 16.20000 7.9555556
[7,] 13.77778 4.9444444
[8,] 13.00000 4.2222222
[9,] 14.60000 6.0444444
[10,] 19.70000 158.9000000
[11,] 16.00000 5.1111111
[12,] 13.90000 2.1000000
[13,] 14.30000 1.5666667
[14,] 14.90000 2.9888889
[15,] 15.30000 5.7888889
[16,] 16.40000 34.4888889
[17,] 15.00000 2.4444444
[18,] 15.00000 3.5555556
[19,] 15.10000 4.9888889
[20,] 14.70000 9.3444444
[21,] 15.20000 10.1777778
[22,] 14.70000 5.1222222
[23,] 17.70000 57.5666667
[24,] 16.50000 6.0555556
[25,] 14.11111 10.6111111
[26,] 13.70000 6.0111111
[27,] 13.50000 2.5000000
[28,] 17.40000 57.1555556
[29,] 13.50000 2.5000000
[30,] 14.30000 2.6777778
[31,] 14.80000 7.7333333
[32,] 14.00000 4.4444444
[33,] 15.30000 14.9000000
[34,] 17.10000 14.9888889
[35,] 14.50000 4.2777778
[36,] 15.70000 14.0111111
[37,] 13.40000 5.1555556
[38,] 15.20000 2.4000000
[39,] 14.50000 3.8333333
[40,] 13.30000 5.1222222
[41,] 14.60000 4.4888889
[42,] 19.90000 174.1000000
[43,] 15.20000 4.4000000
[44,] 13.60000 9.8222222
[45,] 15.10000 2.9888889
[46,] 14.60000 3.3777778
[47,] 14.50000 12.7222222
[48,] 16.10000 6.3222222
[49,] 17.60000 134.9333333
[50,] 15.33333 12.0000000
[51,] 13.10000 6.3222222
[52,] 14.70000 1.3444444
[53,] 15.60000 5.6000000
[54,] 15.00000 1.3333333
[55,] 17.70000 115.7888889
[56,] 15.10000 4.7666667
[57,] 13.70000 3.7888889
[58,] 15.20000 2.4000000
[59,] 14.44444 4.0277778
[60,] 14.90000 6.5444444
[61,] 15.10000 1.4333333
[62,] 15.30000 32.2333333
[63,] 14.60000 4.2666667
[64,] 14.80000 5.0666667
[65,] 13.80000 12.1777778
[66,] 15.20000 9.2888889
[67,] 15.40000 8.0444444
[68,] 14.20000 2.4000000
[69,] 14.22222 4.9444444
[70,] 15.00000 2.8888889
[71,] 21.60000 535.1555556
[72,] 15.40000 5.1555556
[73,] 15.00000 14.6666667
[74,] 14.80000 5.7333333
[75,] 14.20000 7.7333333
[76,] 14.30000 1.5666667
[77,] 14.50000 17.6111111
[78,] 14.70000 8.4555556
[79,] 13.90000 0.9888889
[80,] 16.40000 20.0444444
[81,] 14.50000 2.5000000
[82,] 15.10000 2.5444444
[83,] 15.60000 6.4888889
[84,] 17.50000 61.6111111
[85,] 14.70000 6.4555556
[86,] 14.60000 4.0444444
[87,] 14.50000 1.1666667
[88,] 14.50000 1.6111111
[89,] 16.30000 7.7888889
[90,] 14.70000 1.7888889
[91,] 15.10000 4.1000000
[92,] 15.40000 6.0444444
[93,] 14.10000 2.3222222
[94,] 14.00000 3.1111111
[95,] 14.20000 1.9555556
[96,] 15.00000 2.2222222
[97,] 14.60000 6.7111111
[98,] 14.90000 6.9888889
[99,] 14.10000 2.9888889
[100,] 15.50000 12.2777778I had to trap for cases where the population had gone extinct by gen 6.
Make a plot.
rep_spread_stats <- as.data.frame(rep_spread_stats)
maxd_data <- pull(subset(LerC_spread, Gap == "0p" & Generation == 6), Furthest)
ggplot(rep_spread_stats, aes(x = Mean)) + geom_histogram() +
geom_vline(colour = "red", xintercept = mean(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.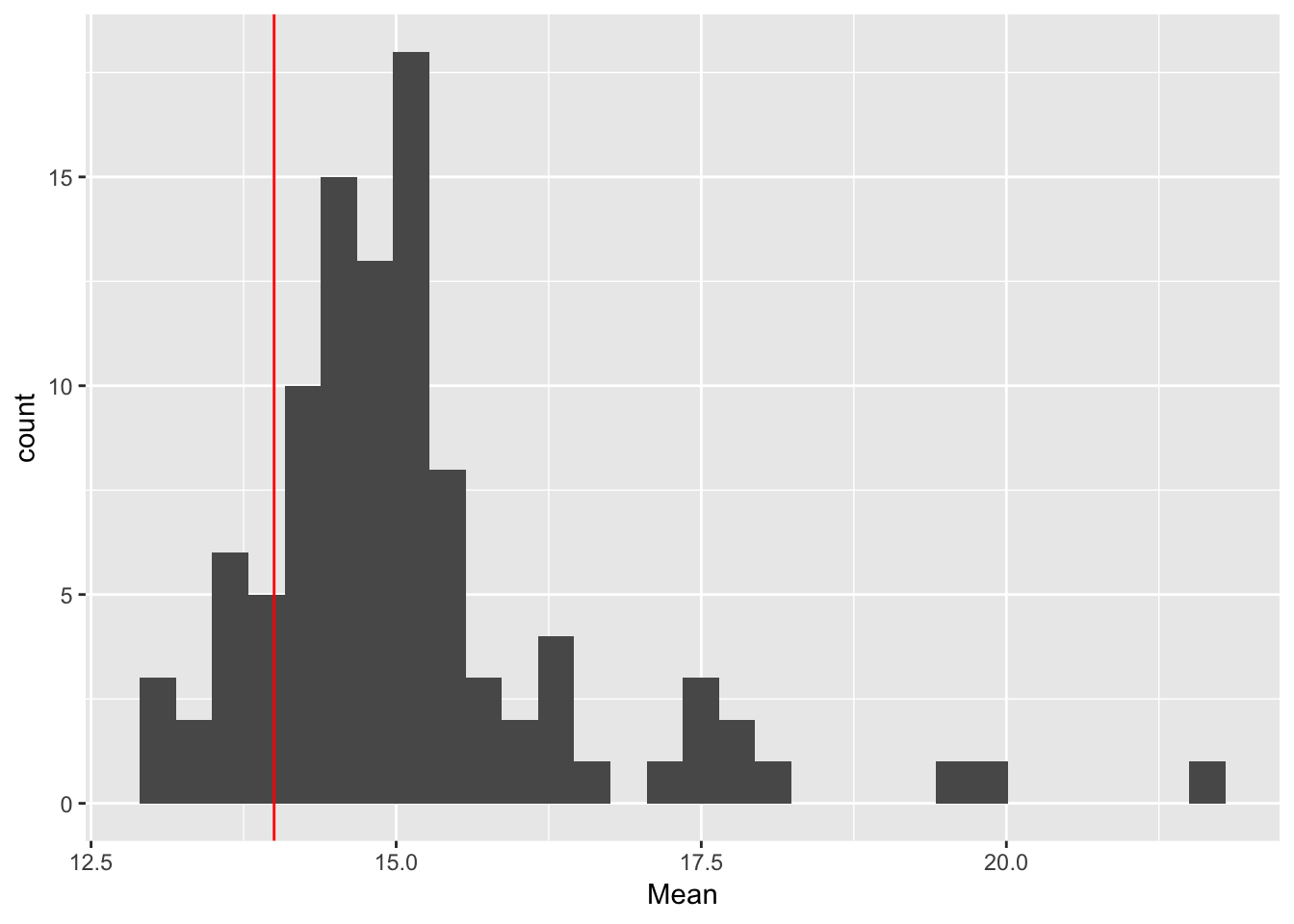
ggplot(rep_spread_stats, aes(x = Variance)) + geom_histogram() +
geom_vline(colour = "red", xintercept = var(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.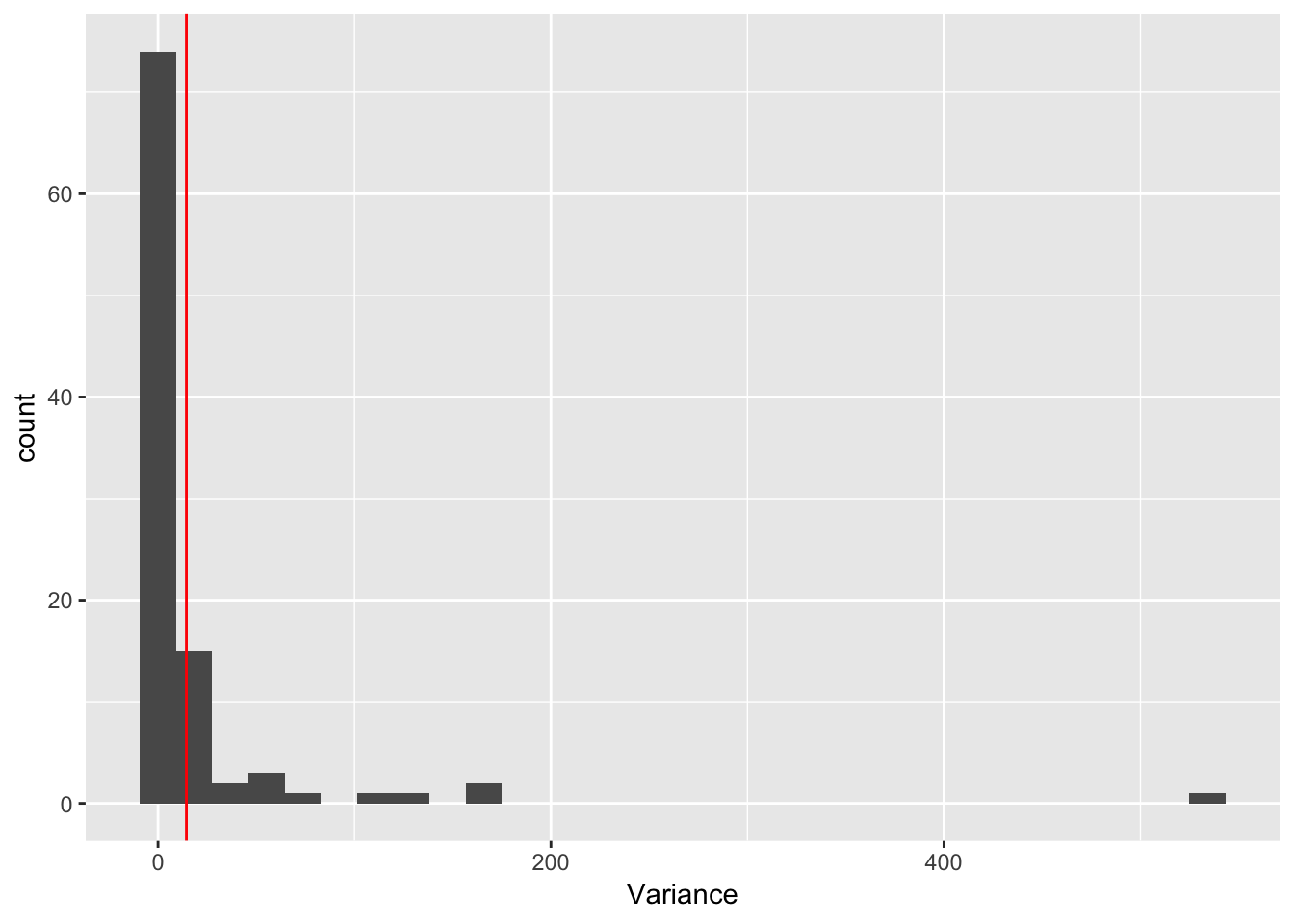 So, the distibution of variances is really skewed and there are some really large values. but the data are right in there. Here is the data: 14, 14.4444444. And here is the mean of the simulations: 15.0927778, 19.5081111. If I trim the most extreme values we get 15.0477324, 14.4353741
So, the distibution of variances is really skewed and there are some really large values. but the data are right in there. Here is the data: 14, 14.4444444. And here is the mean of the simulations: 15.0927778, 19.5081111. If I trim the most extreme values we get 15.0477324, 14.4353741
Just for grins, let’s see what happens if we turn off kernel stochasticity.
nruns <- 100
controls$kernel_stoch <- FALSE
rep_spread_stats <- t(replicate(nruns, sim_mean_var(), simplify = TRUE))Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Infrep_spread_stats <- as.data.frame(rep_spread_stats)
ggplot(rep_spread_stats, aes(x = Mean)) + geom_histogram() +
geom_vline(colour = "red", xintercept = mean(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.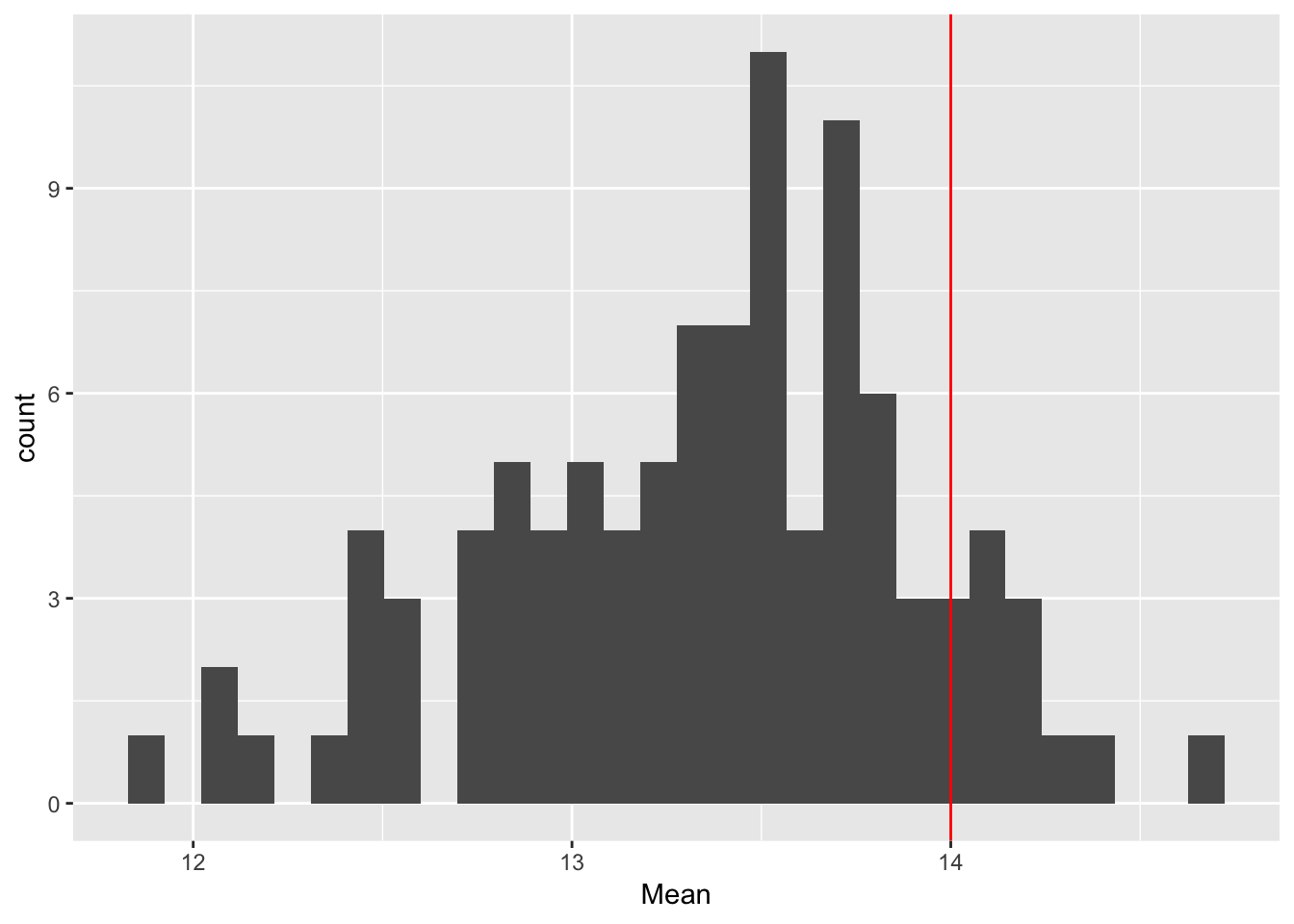
ggplot(rep_spread_stats, aes(x = Variance)) + geom_histogram() +
geom_vline(colour = "red", xintercept = var(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.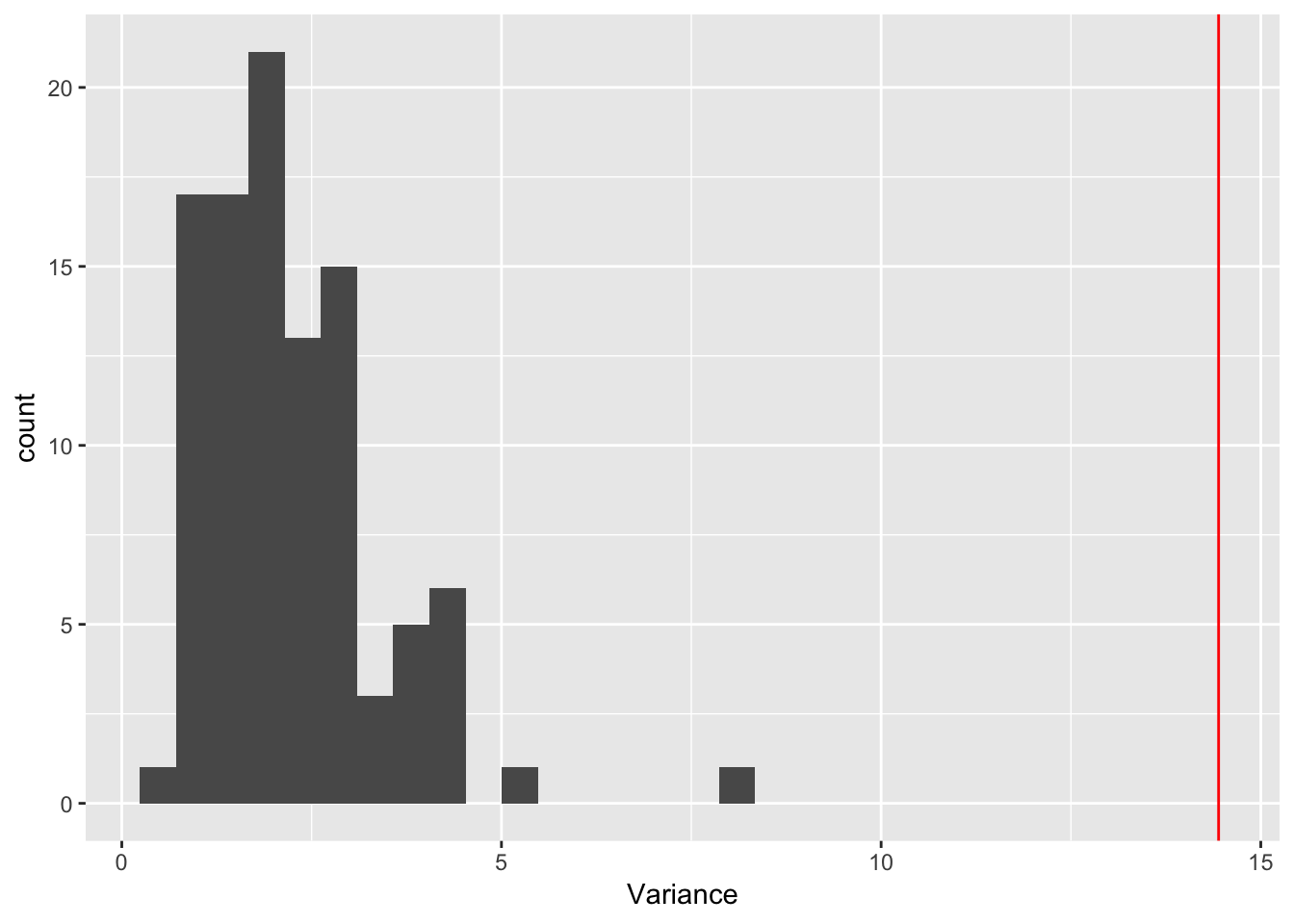 That shoots it all to heck! Let’s try shutting off the others one at a time.
That shoots it all to heck! Let’s try shutting off the others one at a time.
No seed sampling:
nruns <- 100
controls$kernel_stoch <- TRUE
controls$seed_sampling <- FALSE
rep_spread_stats <- t(replicate(nruns, sim_mean_var(), simplify = TRUE))Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Inf
Warning in max((1:length(x))[x > 0]): no non-missing arguments to max;
returning -Infrep_spread_stats <- as.data.frame(rep_spread_stats)
ggplot(rep_spread_stats, aes(x = Mean)) + geom_histogram() +
geom_vline(colour = "red", xintercept = mean(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
ggplot(rep_spread_stats, aes(x = Variance)) + geom_histogram() +
geom_vline(colour = "red", xintercept = var(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.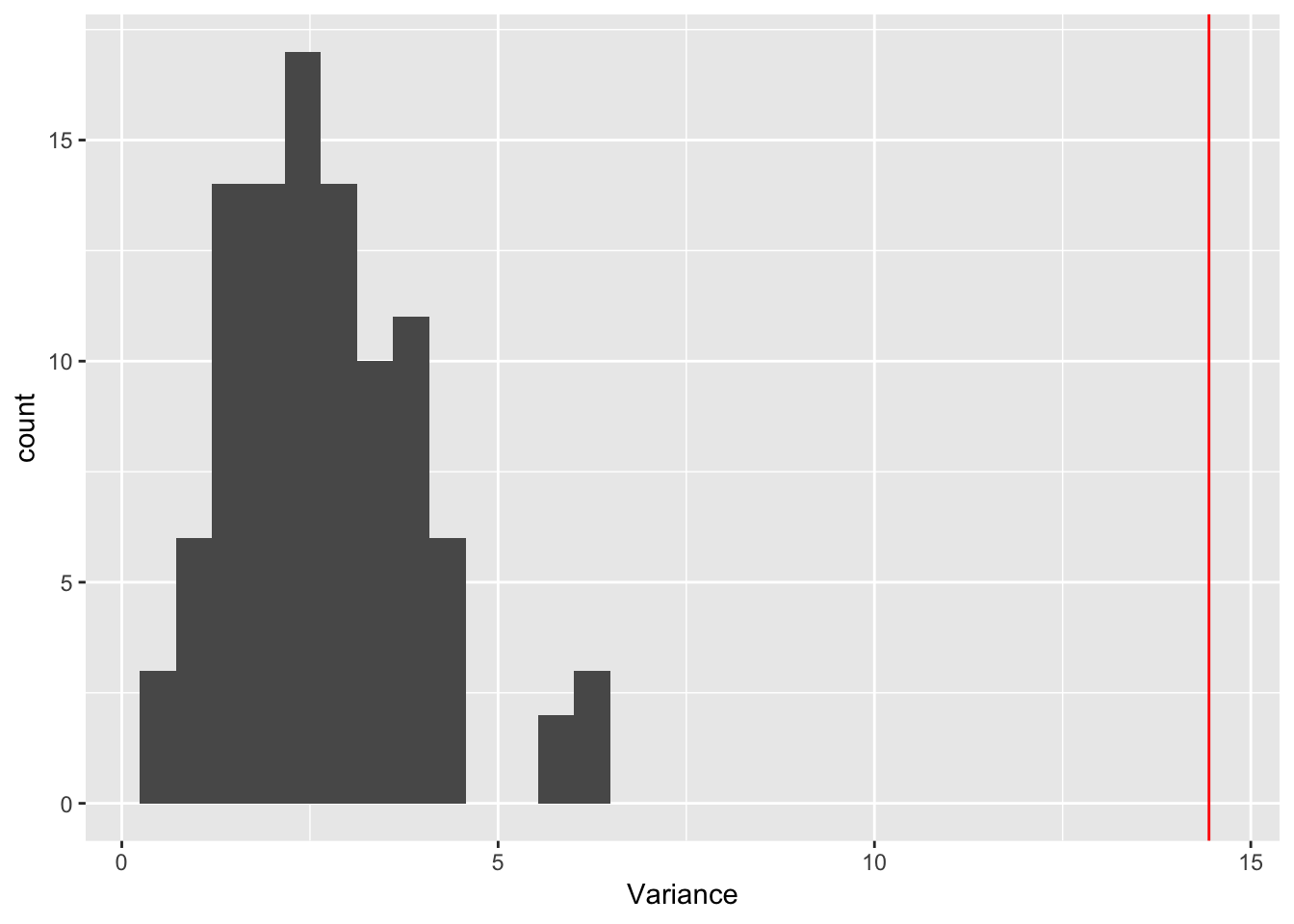
No ES:
nruns <- 100
controls$ES_seeds <- FALSE
controls$seed_sampling <- TRUE
rep_spread_stats <- t(replicate(nruns, sim_mean_var(), simplify = TRUE))rep_spread_stats <- as.data.frame(rep_spread_stats)
ggplot(rep_spread_stats, aes(x = Mean)) + geom_histogram() +
geom_vline(colour = "red", xintercept = mean(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.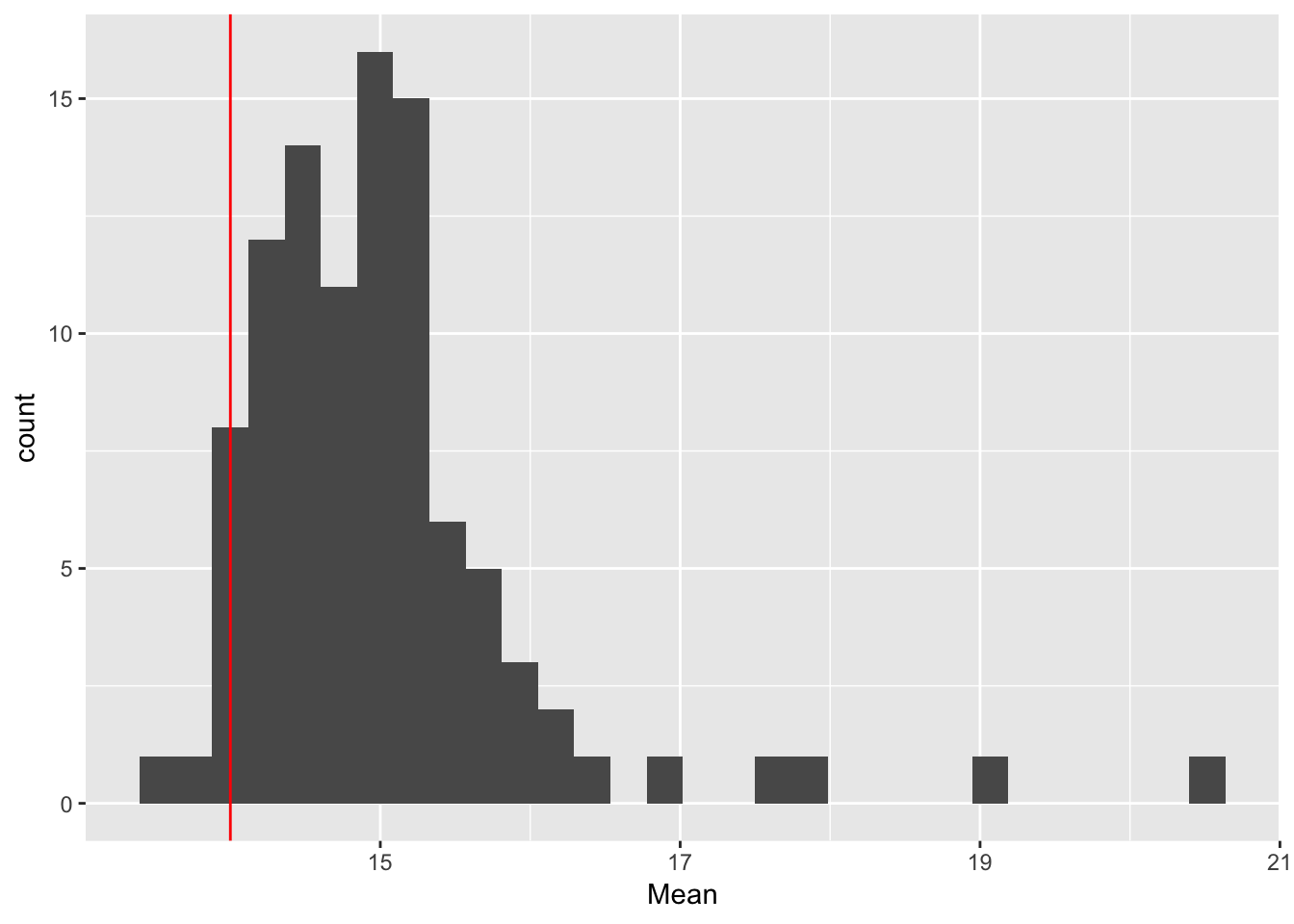
ggplot(rep_spread_stats, aes(x = Variance)) + geom_histogram() +
geom_vline(colour = "red", xintercept = var(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.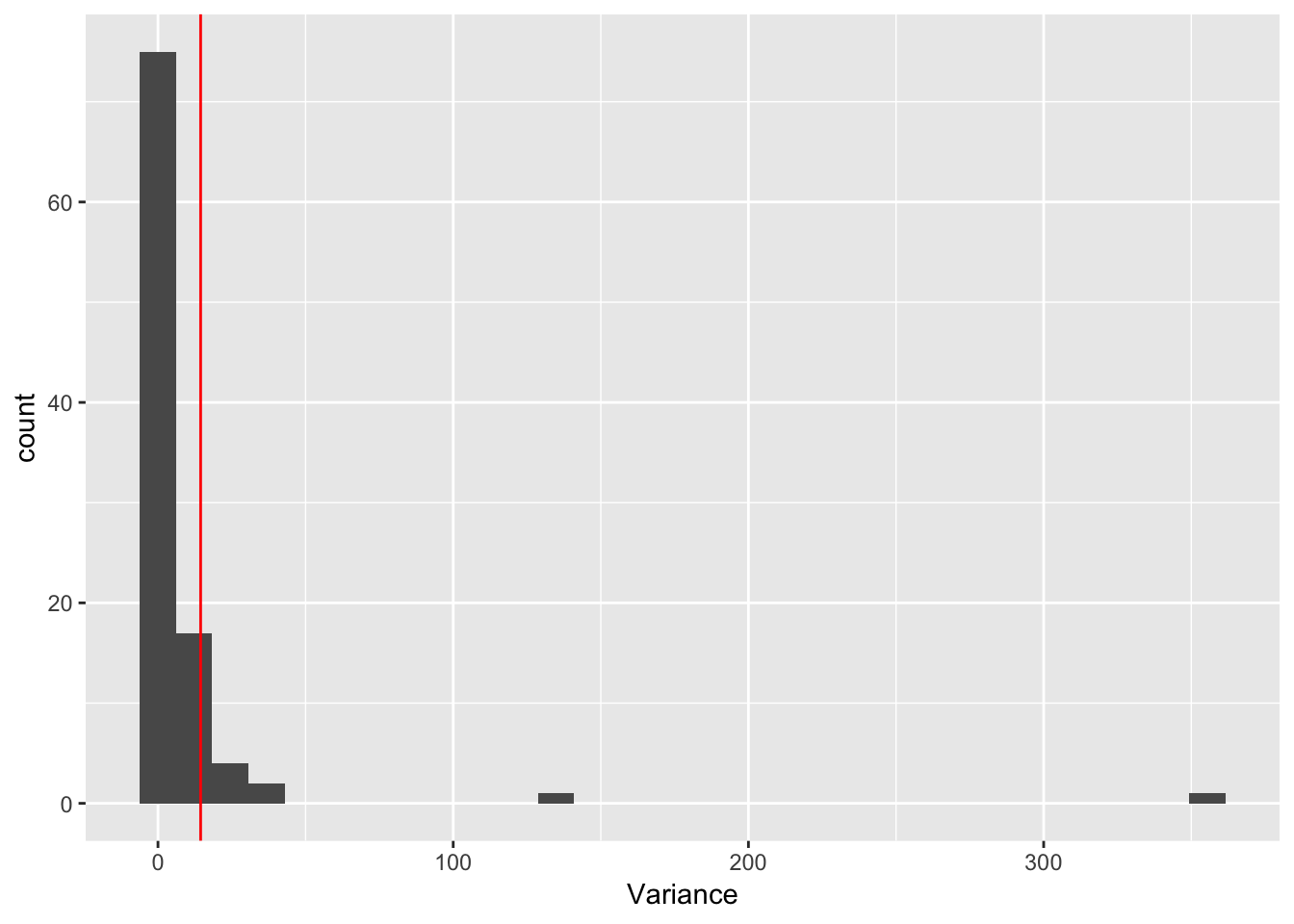
No DS:
nruns <- 100
controls$DS_seeds <- FALSE
controls$ES_seeds <- TRUE
rep_spread_stats <- t(replicate(nruns, sim_mean_var(), simplify = TRUE))rep_spread_stats <- as.data.frame(rep_spread_stats)
ggplot(rep_spread_stats, aes(x = Mean)) + geom_histogram() +
geom_vline(colour = "red", xintercept = mean(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.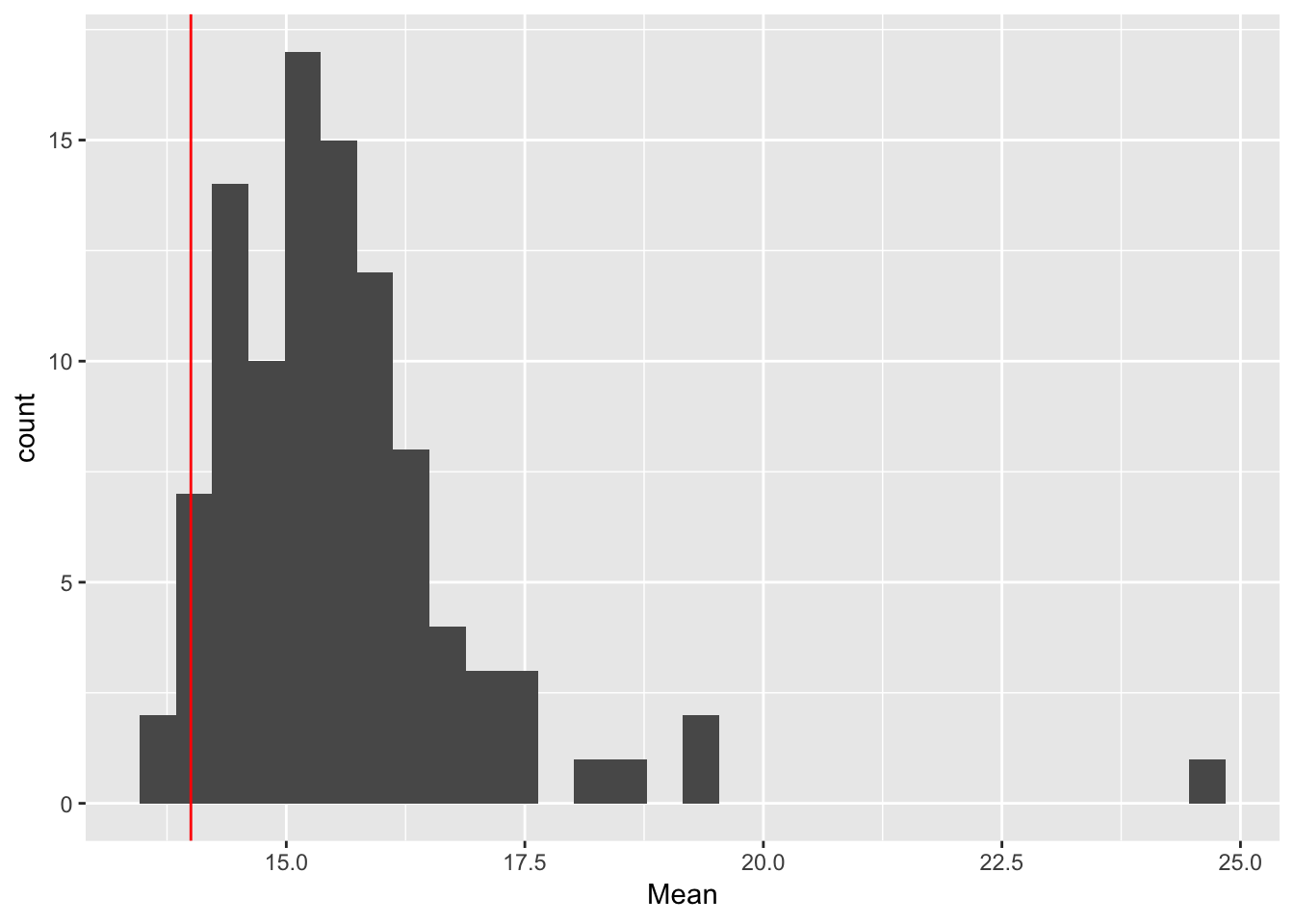
ggplot(rep_spread_stats, aes(x = Variance)) + geom_histogram() +
geom_vline(colour = "red", xintercept = var(maxd_data))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.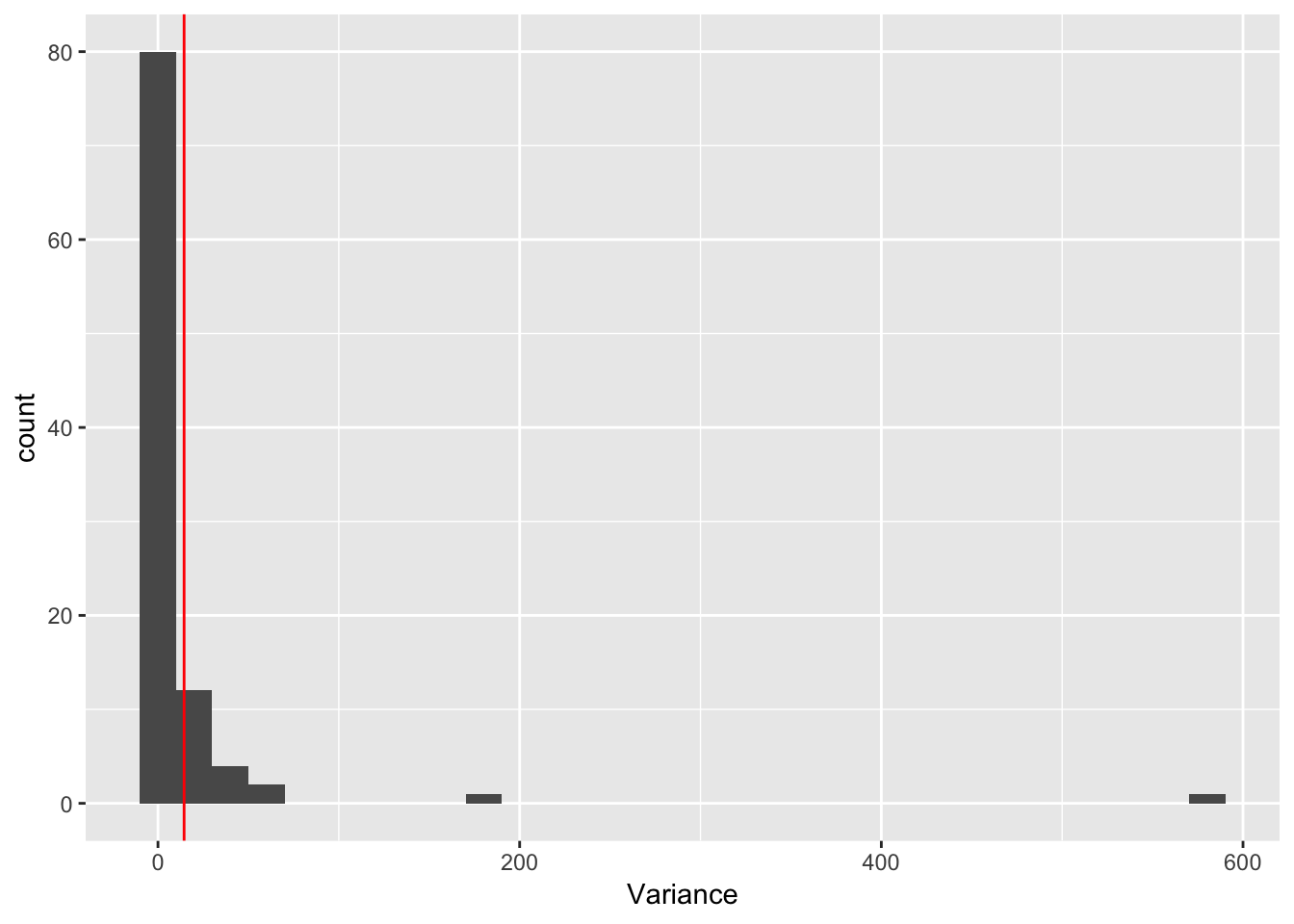
Bottom line conclusions:
- Kernel stochasticity increases the mean spread rate
- Both kernel stochasticity and seed sampling greatly increases the variance in spread rate
- Both ES and DS in seed production decrease the mean spread rate, but have little impact on the variance.